Barcoded magnetic beads are immunomagnetic beads combined with “digital barcodes” say oligonucleotides to create a new, bio-inspired magnetic beads.
How to synthesize barcoded beads used in Microwell Seq?
- Get immunomagnetic beads: carboxyl functionalized magnetic beads from EPRUI, which is in charge of overseas business of Suzhou Knowledge & Benefit Sphere Tech.
- The barcoded oligonucleotides on the surface of the immunomagnetic beads were synthesized by three rounds of split-pool. All the sequences used are listed in supplementary information below.
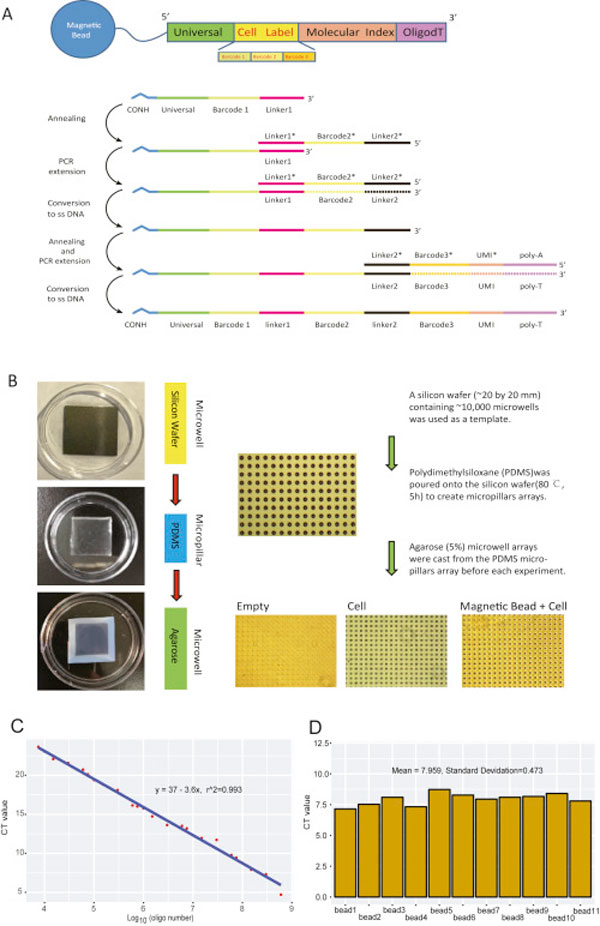
For each batch of bead synthesis, 300-350 μL of carboxyl magnetic beads (50 mg/ml) were washed twice with 0.1 M MES (2-[N-morpholino]ethanesulfonic acid). The beads were then suspended in 0.1 M MES at a final volume of 635 μl. 3.08 mg of EDC (1-ethyl-3 (−3-dimethylaminopropyl) carbomiimide hydrochloride) were added to the beads. 6.2 μL of beads were then distributed into each well of a 96-well plate. 2.5 μL of amino modified oligonucleotide (50uM in 0.1 M MES) were then added into every well. After vortexing and incubation for 20 minutes at ambient temperature, 0.5 μL mix (Add 6 mg of EDC in 100 μL of 0.1 M MES) was distributed into every well. After another round of vortexing and incubation for 20 minutes at ambient temperature, 0.5 μL more mix (Add 6 mg of EDC in 100 μL of 0.1 M MES) was distributed into every well. After vortexing and incubate for 80 minutes at ambient temperature, the beads were collected in 1 mL of 0.1M PBS containing 0.02% Tween-20. After centrifugation, supernatant was removed carefully. The beads were then washed two times in 1 mL of TE (pH 8.0).
In the second split-pool, the beads were washed with water and split into each well of another 96 well plate containing the PCR mix: 1 × Phanta Master Mix (Vazyme) and 5 μM oligonucleotides. The oligonucleotides in every tube encoded a sequence that was reverse complementary to linker 1, a unique barcode and a linker 2 sequence. PCR program was as follows: 94°C 5 min; 5 cycles of 94°C 15 s, 48.8°C 4 min, and 72°C 4 min; 4°C hold.
In the third split-pool procedure was the same as the second one. PCR program was as follows: 94°C for 5 min, 48.8°C for 20 min, 72°C for 4 min and 4°C hold. The oligonucleotides used in every tube encoded a linker 2 reverse complementary sequence, a unique barcode, a UMI sequence and a poly T tail. Beads were resuspended in 1 mL of ddH2O. To remove complementary chains, put beads into 95°C water bath for 6 min, separate beads with magnetic separator and remove the supernatant quickly for 2 times. Beads could be stored in TE-TW (10 mM Tris pH 8.0, 1 mM EDTA, 0.01% Tween20) for 4 weeks at 4°C.
