A group of scientists from ZheJiang University have successfully mapped the mouse cell altas by Microwell-seq in February 2018. They delveloped a new Microwell-seq, a high-throughput and low-cost scRNA-seq platform using simple and inexpensive devices to map the mouse cell altas for the first time in human history. By using the new device, the cost of sequencing library generation for a single cell is estimated to be reduced to USD$0.2 while it was around USD$15 per cell in the past.
(Professor Guoji Guo’s Research Group who is in the middle of the first row)
scRNA-seq or Single-cell RNA sequencing technologies are poised to reshape the current cell-type classification system. However, a transcriptome-based single-cell atlas has not been achieved for complex mammalian systems. Using Microwell-seq, the scientists in Zhejiang University leading by Professor Guoji Guo, analyzed more than 400,000 single cells covering all of the major mouse organs and constructed a basic scheme for a mouse cell atlas (MCA). They reveal a single-cell hierarchy for many tissues that have not been well characterized previously. They even built a web-based “single-cell MCA analysis” pipeline that accurately defines cell types based on single-cell digital expression. Their study demonstrates the wide applicability of the Microwell-seq technology and MCA resource.
Microwell-seq, a simple method to profile thousands of single cells by transcriptome analysis utilizing an agarose-constructed microwell array and barcoded beads. Microwell-seq has advantages in convenience and simplicity, which should make the method widely accessible. Using Microwell-seq, the scientists constructed a first stage “mouse cell atlas” with more than 400 k single-cell transcriptomic profiles from 51 mouse tissues, organs, and cell cultures. The resulting map covers more than 800 major cell types and potentially more than 1,000 cell subtypes in the mouse system. We integrated published high-throughput single-cell data and built the MCA website database for the scientific community. Finally, they constructed a single-cell MCA (scMCA)” tool that accurately defines cell types based on single-cell digital expression. Future efforts in data accumulation and integration will eventually lead to creation of a comprehensive mammalian cell map that will facilitate related basic research and clinical applications.
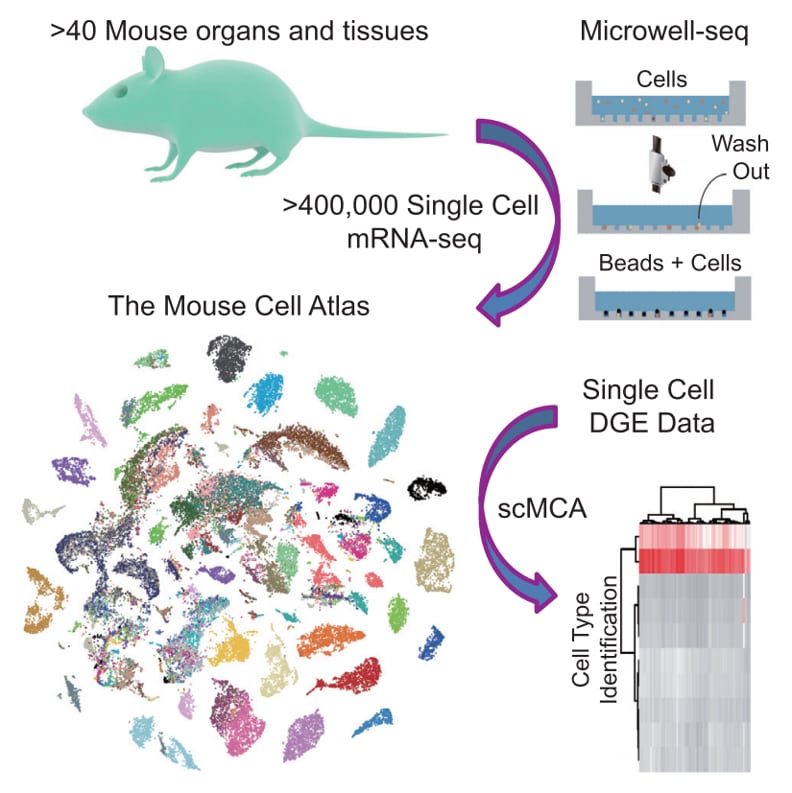
(Mouse Cell Altas)
EPRUI is able to supply the barcoded 20-25um carboxylated magnetic silica beads used in the experiment. Kindly contact sales@epruibiotech.com for more details.
References:
- Mapping the Mouse Cell Atlas by Microwell-Seq https://doi.org/10.1016/j.cell.2018.02.001
- single-cell MCA analysis Website: http://bis.zju.edu.cn/MCA/
